
Guidelines
These guidelines are introduced in 2023 (PMID: 37426048) and are based on our collective experience of building representations of disease mechanisms, as well as on the descriptions of the earlier published disease maps and comprehensive pathway maps. In each case, a disease map is designed while taking into account the goals and objectives of a particular project, specifics of disease conditions and disease subtypes.
There are training materials, definitions, templates and examples for different stages of the disease map development:
Map planning template
Learning SBGN
Knowledge formalisation
Annotation
Designing map architecture
Glossary
Disease map development workflow
The development workflow can be subdivided into nine steps within four phases: planning, curation, publishing and application.
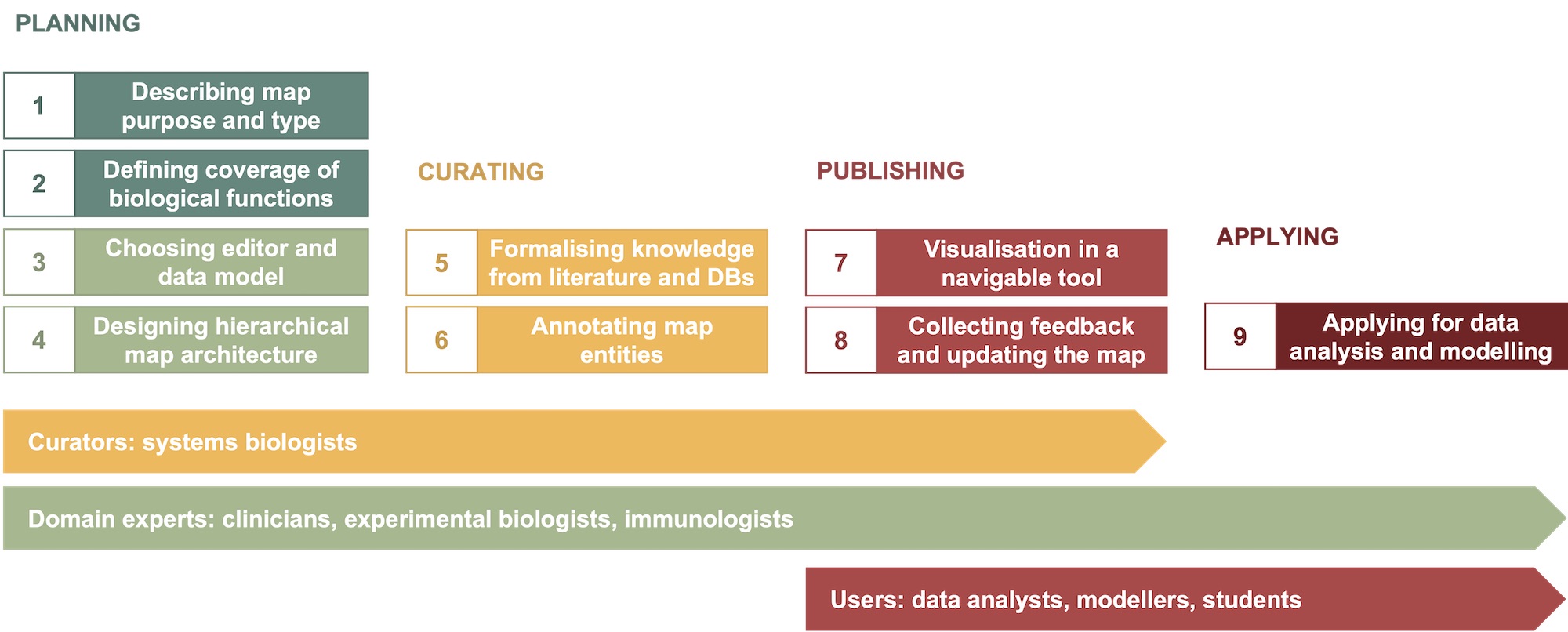 Figure 1. The map development workflow.
Figure 1. The map development workflow.